Abstract
DNA nanotechnology can be leveraged to engineer nanoscale biochemical reactions, and thus, revolutionize biomanufacturing. The programmability is encoded in the interactions between base pairs of the nucleic acids. Functional nanostructures can be envisioned and formed, such as DNA nanostars, whose properties can be fine-tuned by engineering the number of arms or base pairs per arm and can yield synthetic condensate structures, and DNA-based enzymes that exhibit peroxidase-like activity. For example, certain guanine-rich sequences of DNA can fold into a quadruplex structure, bind a hemin co-factor, and catalyze a peroxidation reaction in which the substrate ABTS (2,2’-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid)) gets oxidized by hydrogen peroxide and results in a colorimetric change. Because ABTS produces a blue-green color change upon oxidation, it can be used to visually observe the peroxidation reaction taking place within the DNA condensates. In this work, peroxidase-mimicking DNAzymes were used to catalyze colorimetric peroxidation within DNA condensate compartments; and toehold-mediated strand displacement (TMSD) was explored as a strategy to program the peroxidation reaction–specifically, by unwinding the G-quadruplex structure, which would effectively turn the reaction “off”. TMSD is a method of designing a single strand of DNA with an additional overhang region, called a toehold, to oust and replace a second strand attached to the toehold-possessing target strand. The presence of complementary toeholds on both the invading strand and the target strand increases the thermodynamic probability of displacing the single DNA strand originally bound to the target. Here, TMSD was adapted for use in ‘turning off’ the DNAzyme-catalyzed peroxidation reaction, either by preventing folding or disrupting the folded structure of the DNAzyme. A displacer strand complementary to the DNAzyme/toehold region was designed and added to the reaction mixture at different time points and concentrations for this purpose. Elucidating mechanisms to unwind the G-quadruplex structure of DNAzymes has promise in treating genetic disorders caused by unregulated G4 formation in the human genome. Furthermore, DNA nanotechnology can be used to compartmentalize, functionalize, and program the release of bioactive molecules in drug delivery strategies and other synthetic biology applications, highlighting the potential of TMSD to program DNA-based bioreactors. This high impact study, carried out as part of the NSF Future Manufacturing program at Pasadena City College in collaboration with UCLA, UCSB, and Caltech, allowed undergraduate researchers to design and conduct their own experiments within a community college setting after undergoing scientific training by graduate students and postdocs from our collaborators’ institutions. It also provided opportunities to communicate the scientific research through writing, poster presentations at national conferences, and teaching in courses and STEM outreach. The student researchers of the PCC nanostar program applied their knowledge in a classroom setting, where they taught other undergraduate students how to conduct aspects of this research in a General, Organic and Biochemistry laboratory course at PCC. This article underscores the importance of creating significant research and teaching opportunities for students as they begin their careers in STEM, impactful mentorship through undergraduate research, and the creativity involved in modern synthetic biology research and in the development of accessible and innovative science lessons.
Keywords: undergraduate research; DNA nanotechnology; Synthetic Biology; General, Organic & Biochemistry; scientific method; workforce development; DNA nanostar; equity in STEM; STEM education; STEM outreach; women in science
© 2025 under the terms of the J ATE Open Access Publishing Agreement
Introduction
The ability to associate, separate, sort and more generally, spatially organize components is central to manufacturing. These tasks have been mastered in industrial biochemical processes but remain a challenge to embed in biochemical reactors at the micro- and nanoscale. Toward the aim of engineering nanoscale reactors using DNA nanotechnology [1],[2], we take our inspiration from biology, as cells have evolved machinery to autonomously segregate DNA, proteins, small molecules, and entire reaction pathways [3], [4],[5]; and there is growing evidence that liquid-liquid phase separation is a key phenomenon enabling cells to control molecules in space and time [6],[7],[8],[9]. Droplet-like organelles spontaneously condense within cells and host specific molecules and reaction pathways. Oftentimes these structures have multiple internal compartments or layers by which they partition biochemical reactions. Achieving the power of cellular condensates with synthetic materials [10],[11] has the potential to revolutionize biomanufacturing by facilitating engineering of microreactors that can organize molecules and their reactions biochemically.
DNA nanotechnology can be used to engineer synthetic condensates [12],[13],[14],[15],[16],[17]; the programmability of DNA is inherent in its structure and interactions between base pairs. For example, the sequence of base pairs determines the folding, interaction strength, binding to target molecules, triggered release of target molecules, sensing of other molecules and even nontrivial chemical reactivity (i.e. catalysis by DNAzymes). DNA nanotechnology has demonstrated fine control over the thermodynamic and kinetic interactions of DNA or RNA strands [18]. Thus, association and dissociation of DNA or RNA monomers can be easily programmed, and further, it is possible to modulate the kinetics of condensation and phase transition through established methods of strand displacement [18],[19].
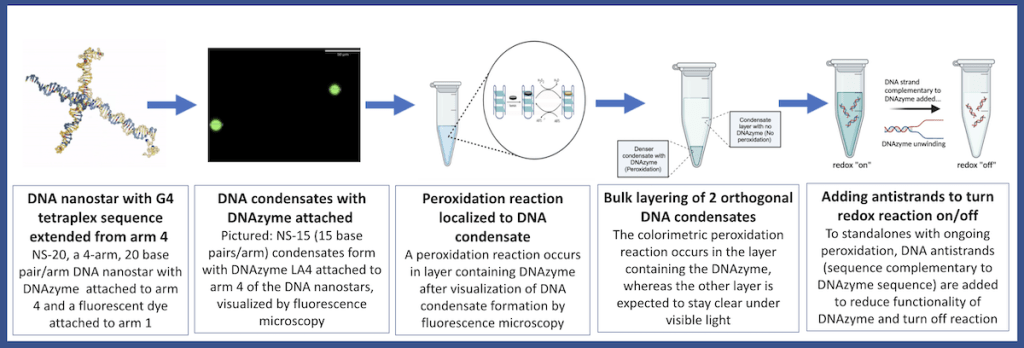
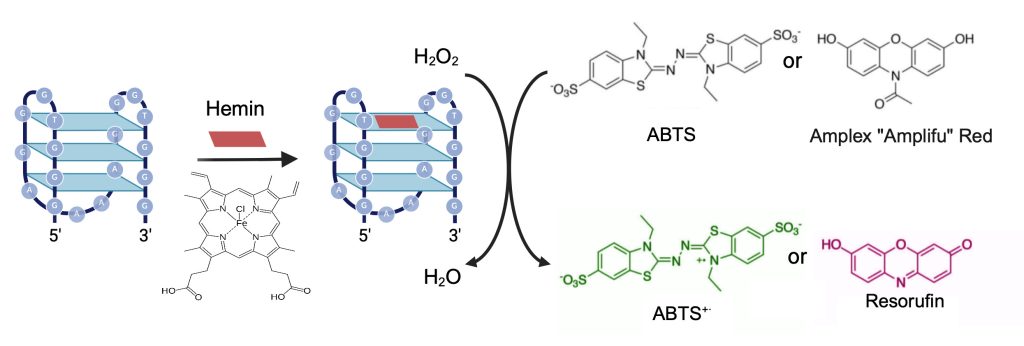
Through the NSF Future Manufacturing Research Program at Pasadena City College (PCC), we train and mentor a creative and resilient manufacturing workforce [20] as they engage with emerging nucleic acid nanotechnologies. The overall aim of the research conducted through this undergraduate research program is to use the methods of DNA nanotechnology to engineer programmable DNA-based biochemical reactions localized to synthetic DNA condensates. In our envisioned strategy, shown in Figure 1, 4-armed DNA nanostars [21],[22] with either 15, 20, or 25 base pairs per arm are designed with 6 base-pair sticky ends, which allow the annealed nanostars to form condensates [23], and a fluorophore on one of the arms for visualization. A different arm of the DNA nanostar is extended with the sequence for a DNAzyme [24],[25],[26],[27],[28],[29], a G4 quadruplex that binds hemin and exhibits peroxidase-like activity. Previously, PCC undergraduate researchers tested various parameters to achieve localization of DNAzyme-mediated peroxidation on the microscale, first screening a panel of DNAzymes for peroxidase activity, selecting DNAzymes for integration into condensates, and testing two different redox substrates, ABTS (2,2’-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid)) and Amplifu Red, which become oxidized by hydrogen peroxide and result in a colorimetric response (Figure 2). This color change allows researchers to correlate blue/pink colored condensates to fluorescent condensates to confirm localization through microscopy [30]. Notably, the PCC nanostar researchers ran molecular simulations of the designed DNA nanostars using oxDNA to inform their research and education efforts [31]. To achieve programmability, the next research aim was to use strand displacement to ‘turn off’ the DNAzyme-catalyzed reaction.
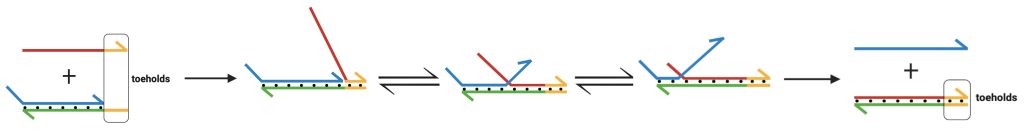
Building upon previous work [30],[31], the next cohort of PCC nanostar undergraduate researchers set out to explore toehold-mediated strand displacement (TMSD) [32],[33] as a means to program the DNAzyme-catalyzed peroxidation reaction localized to synthetic DNA condensates. Peroxidase-mimicking DNAzymes were formed and used to catalyze colorimetric peroxidation within DNA condensate ‘compartments’; and TMSD was adapted as a strategy to program the peroxidation reaction. TMSD is a method of designing a single strand of DNA with an additional overhang region, called a toehold, to oust and replace a second strand attached to the toehold-possessing target strand (Figure 3). The presence of complementary toeholds on both the invading strand and the target strand increases the thermodynamic probability of displacing the single DNA strand originally bound to the target [33]. The process of toehold-mediated strand displacement allows control over the rate at which various reaction pathways occur [19]. We hypothesized that TMSD could be used to ‘unwind’ the G quadruplex structure [34], which would effectively turn the peroxidation reaction off. Thus, in this work, TMSD was employed to turn off the DNAzyme-catalyzed peroxidation reaction, either by preventing folding or disrupting the folded structure of the DNAzyme. A displacer strand complementary to the DNAzyme/toehold region was designed and added to the reaction mixture at different time points and concentrations for this purpose.
DNAzyme-catalyzed peroxidation has been carried out in hydrogel beads [36] and RNA catalysis has been confined within membraneless organelles [37]. This research project expands upon previously established methods [31],[32] to localize DNAzyme-catalyzed peroxidation within microscopic condensates of a four-arm DNA nanostar condensate layer and explores methods to program the reaction. Specifically, this study aims to ‘turn off’ the reaction using TMSD, representing a novel approach toward the development of nanoscale bioreactors using nucleic acids. DNA sequences complementary to the DNAzyme were designed and toeholds were incorporated on the invading and target strands, reasoning that the toeholds would facilitate the displacement reaction, unwind the G4 quadruplex, and reduce the activity of the DNAzyme. The overall goal of this research was to evaluate the programmability of DNAzyme-mediated reactions within synthetic DNA condensates using displacer strands complementary to the DNAzyme sequence. By utilizing TMSD, we expected to suppress the effect of the DNAzyme by unwinding or preventing the folding of the G-quadruplex. G4s are a ubiquitous feature in the human genome that play a role in gene expression. However, when G4 formation is not adequately regulated in vivo due to a deficiency in helicase activity, its accumulation can threaten genetic stability, resulting in genetic disorders and cancers. Therefore, a mechanism to unwind the G-quadruplex that could be translated to in vivo applications has the potential to yield revolutionary advances in genetic disease therapies [38].
This research was carried out through the Future Manufacturing undergraduate research program at PCC, which has empowered many underrepresented students through scientific research, thus contributing to diversity in the STEM workforce. Five female undergraduate students at PCC were selected for the 2024 summer cohort, and they began the program with a research techniques bootcamp in UCLA’s Dynamic Nucleic Acid Systems laboratory, where they were taught by graduate students, postdocs and professors at the collaborating institutions of UCLA, UCSB, and Caltech. Here, the undergraduate researchers were trained in methods they would be using in their own research, while also experiencing the type of research and environment they could expect if they chose to pursue a research career in the future. Following the research techniques bootcamp, students returned to PCC to perform their own experiments for the next six weeks, 20 hours per week, with the goal of localizing biochemical reactions to DNA condensates and strand displacement to program these reactions within the condensates. As part of this transformative experience, students were mentored as they conducted their research; they were provided opportunities to write about their research results, present their findings at the PCC Natural Sciences Division Poster session [39],[40] and the American Chemical Society National Meeting in 2025, and share their experience as a nanostar researcher with peers at the Career and Research Internship Panel at PCC’s Pi Day [30]. They were also given the unique and empowering experience to teach their experimental research methods to a class of undergraduate students in Prof. Jillian L Blatti (Dr. J.B.)’s Chemistry 2B: General, Organic & Biochemistry laboratory course as learning assistants. Throughout the program, students were exposed to and immersed in various aspects of the STEM research field and given the opportunity to explore STEM career paths. They developed invaluable professional skills [41], felt a sense of belonging to the scientific community, and formed a supportive ‘nanostar network’ with inspirational female role models to encourage them through their career trajectories. This research program has inspired underrepresented students in STEM [42], recruited talent from diverse backgrounds, and provided further opportunities for students once they transferred to UCLA. Notably, several PCC nanostars continued research in our collaborators’ laboratories.
Methods
Materials and Reagents
All of the DNA strands used in these experiments were analyzed by NUPACK software [43] and purchased from Integrated DNA Technologies (IDT). After quantifying the absorbances of DNA strands at 260 nm using a Nanodrop One UV-Vis Spectrophotometer, their concentrations were calculated using Beer’s Law and the extinction coefficients provided on the IDT oligonucleotide specification sheets (Table S1). DNA nanostar strands without a G4 or toehold attached were diluted tenfold prior to quantification, and their true concentrations were backcalculated.
In experiments with standalone DNA enzymes, the negative control polyT18 (composed of 18 thymine bases) and the G4 5xG3T were used without any toeholds. The polyT18 negative control does not fold into a quadruplex or bind hemin, and so, should not catalyze the reaction. Further, the 5xG3T-T1-displacer strand was added to tubes with standalone 5xG3T to determine if the displacer strand had any effect on slowing down the peroxidation reaction when it had no toehold with which to facilitate binding.
In experiments with DNA condensates formed from four-armed nanostars with 15 base pairs per arm, when the 5xG3T DNAzyme and toehold were attached to 50% of the nanostars’ fourth arm, the 5xG3T-T1-anti ‘displacer’ strand, which was complementary to the G4 and toehold components of the nanostar, was added to the reaction tubes both prior to and after annealing to analyze the effect of toehold-mediated strand displacement on slowing down the peroxidation reaction.
Buffer Preparation
The DNAzyme-catalyzed peroxidation reaction in this study takes place in 1X reaction buffer, which contains 10 mM Tris-HCl (pH 7.0-7.5), 100 mM KCl, 0.05% (v/v) Triton X-100, and 1% (v/v) DMSO [27]. To form the DNAzyme, to the folded G4, hemin was added (dissolved in DMSO to result in a stock concentration of 5mM). To initiate the reaction, substrates were added to the DNAzyme: ABTS (tablets dissolved in milli-Q water to result in a stock concentration of 18.226 mM) and hydrogen peroxide. These were all diluted daily with milli-Q water and 1X reaction buffer to make fresh solutions of 50 µM hemin, 6 mM ABTS, and 6 mM hydrogen peroxide. All buffer reagents were purchased from Sigma-Aldrich with the exception of KCl, which was purchased from Thermo Fisher Scientific.
Formation and Peroxidation of Standalone G4/Hemin Complexes with Displacer Strands
The standalone G4 strands were diluted in reaction buffer and milli-Q ultrapure water to reach a final concentration of 10 µM in 1X reaction buffer. The strands were then folded into the G4 structure via snap cooling, which entailed heating to 95 °C for ten minutes then cooling immediately in an ice bath for 15 minutes [27]. In a separate tube, the freshly prepared 50 µM hemin solution in 1X reaction buffer was incubated in a mixture of the 10 µM snap-cooled G4 strand and 1X reaction buffer (5 µM final concentration of G4/hemin catalyst) for approximately 1 hour at room temperature while covered. After incubation, in another PCR tube, a 1:1 ratio of the G4/hemin catalyst and 6 mM ABTS in 1X reaction buffer were added along with 70 µL of 1X reaction buffer to result in 90 µL of solution. Lastly, 10 µL of 6 mM hydrogen peroxide in 1X reaction buffer was added to initiate the reaction. The final solution contained 0.5 µM G4/hemin catalyst, 600 µM ABTS, 600 µM hydrogen peroxide, and 1X reaction buffer. The reaction was visualized, photographed using a UHM-350 camera, and quantified by measuring absorbance at 420 nm (absorption maximum of ABTS͘ +) using a Nanodrop One UV-Vis Spectrophotometer every 2 minutes over a 10-minute period.
Formation of NS15 DNA Condensates with G4/Hemin Complexes, Toeholds, and Displacer Strands and Peroxidation
To form microscale condensates, four-arm DNA nanostars with 15 base pairs per arm were designed to include two adenine bases without complements between arm-forming sequences to promote bending to the nanostar shape and to end with a six nucleotide long palindromic sticky end overhang with sequence GCTAGC [30],[31]. These palindromic sticky ends facilitate interactions between nearby nanostars via Watson-Crick base-pairing [21] and promote condensation. One of the arms of the DNA nanostar contains a FAM fluorophore for visualization using fluorescence microscopy, and this represents 5% of strand 1.
The DNA nanostar strands were diluted tenfold and the four strands to form nanostar 15 were mixed in equimolar amounts to achieve a final concentration of 20 µM per strand. In addition to the four strands of nanostar 15, 50% of the total concentration of strand 4 included the G4 5xG3T in the control tubes (the tubes that should turn blue-green to show that the reaction occurred), whereas 50% of the total concentration of strand 4 included the G4 5xG3T with an attached toehold (corresponding to the toehold on the antisense strands) in the tubes where the displacer strand would be added. The displacer strand with a complementary toehold was added at 2X, 5X, and 10X the concentration of strand 4 with the toehold and G4, either before or after annealing. To anneal the nanostars, the solutions were placed in a thermocycler set to 95 °C for ten minutes, then slowly cooled to room temperature at a rate of 1 °C per minute.
To the DNA nanostars with G4s with and without toeholds, two different methods for binding the hemin co-factor with the G-quadruplex were explored: adding the hemin prior to annealing or adding hemin after annealing had been completed. 6 µL of 50 µM hemin in 1X reaction buffer was added directly into the reaction tube and mixed with the other reagents before heating or added directly into the reaction tube with the annealed nanostars and allowed to incubate for approximately 1 hour at room temperature while covered after heating. After this step, a mixture of 10 µL of 6 mM ABTS in 1X reaction buffer and 70 µL of 1X reaction buffer was added to the tube, followed by 10 µL of 6mM hydrogen peroxide in 1X reaction buffer to initiate the reaction. The reaction was visualized and photographed using a UHM-350 camera microscope at 1.5X resolution every 2 minutes over a 10-minute interval.
Visualization of DNA Condensates with DNAzymes, Toeholds, and Displacer Strands
To confirm the formation of DNA condensates upon which the toehold-mediated strand displacement experiments were conducted, their reaction tubes were sampled, dyed with dilute SYBR (gold) dye, and viewed under an Echo Revolve Fluorescence Microscope. The SYBR dye was initially diluted by adding 2.5 µL into 25 mL of Tris-HCl (pH 7.0-7.5) buffer, shaking the solution, and covering it with aluminum foil to protect the light-sensitive dye [44]. Then, 0.5 µL of the diluted dye was added to 5 µL of the condensates. For visualization, 2.5 µL of this mixture was pipetted onto a microscope slide within a Parafilm chamber; condensates were observed under the FITC channel of the fluorescence microscope.
Results and Discussion
Experimental Design
The focus of this study was to program a DNAzyme-catalyzed peroxidation reaction localized to synthetic DNA condensates. As previously mentioned, 4-arm DNA nanostars with 15 base pairs per arm and palindromic 6 base pair sticky ends were designed; a fluorophore was attached to 5% of arm 1 strands and the DNAzyme 5xG3T was extended from 50% of arm 4 strands. It was previously determined that G4 quadruplexes extended from strand 4 can fold under the DNA nanostar annealing conditions, bind hemin, and catalyze peroxidation using hydrogen peroxide and ABTS (or Amplifu Red) substrates [30]. To explore TMSD in programming the DNAzyme-mediated reaction, a complementary displacer strand was designed for the G4/DNAzyme 5xG3T, and it included a toehold to promote strand displacement of the G4 quadruplex [18],[19]. The goal of TMSD was to prevent or slow down the redox reaction by unwinding the G4 quadruplex, which would render it inactive. We hypothesized that by introducing a displacer strand complementary to the G4 sequence, the toeholds at the tail ends of the G4 and displacer strand would bind together and unravel the folded G4 quadruplex, disrupting the hemin co-factor’s ability to bind to the quadruplex, and thus, prevent DNAzyme-catalyzed peroxidation. In the absence of a G4/hemin complex, the peroxidation reaction should not be catalyzed, resulting in little to no blue-green color change.
Before attempting toehold-mediated strand displacement on DNA condensates with toeholds, the displacer strand was added to solutions of standalone 5xG3T DNAzymes without toeholds to determine if it had any effect on slowing down the peroxidation reaction. Then, TMSD was attempted on DNA condensates formed from nanostars containing 5xG3T and a toehold on strand 4 to explore its ability to turn off the DNAzyme-catalyzed peroxidation reaction on the microscale using complementary displacer strands with toeholds. This displacer strand was added to the reaction tube at 2X, 5X, and 10X concentrations—compared to the concentration of strand 4/DNAzyme with a toehold—either before or after annealing to prevent or stop peroxidation. The hemin co-factor was also added to the reaction mixture either before or after the annealing process, to determine if the timing of addition impacts whether the G4/hemin complex will form, and thus, whether the peroxidation reaction will be disrupted in the presence of the displacer strands.
Strand Displacement of Standalone DNAzymes without Toeholds
As the use of toehold-mediated strand displacement to disrupt the catalytic ability of a G4/hemin complex has not been previously studied, the process of strand displacement was attempted first on standalone DNAzymes without the presence of toeholds or condensates. The assumption was that the displacer strand may have some ability to attach to and disrupt the folded G4 structure even without facilitation of a toehold with which to bind.
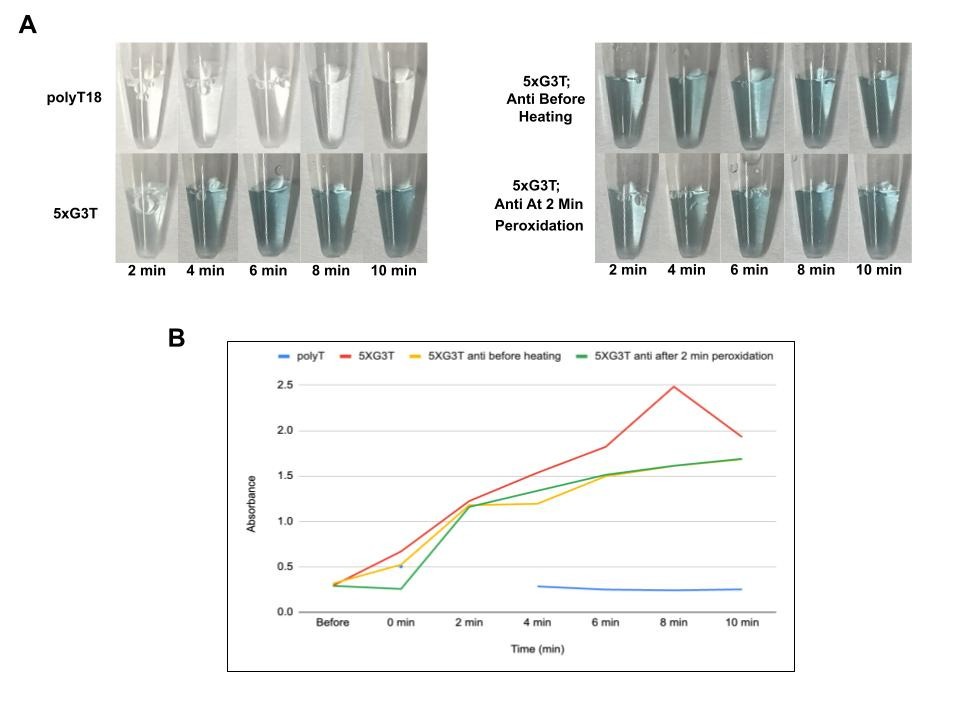
Predictably, the polyT18 negative control solution stayed clear. The reaction tube consisting of standalone 5xG3T provided a baseline blue color to which to compare the tubes where displacer strands would be added (Figure 4A). Visually, the tubes in which displacer strands were added prior to snap cooling and after 2 minutes of peroxidation both looked to turn as blue or slightly less blue than the standalone 5xG3T tube (Figure 4A); however, quantitatively, the absorbance values of these two tubes (at the absorption maximum of ABTS͘ + at 420 nm) plateaued 6-10 minutes post peroxidation in comparison to the positive control (Figure 4B). This suggests that the displacer strand is somewhat successful in slowing down the peroxidation reaction even without the presence of a toehold.
Toehold-Mediated Strand Displacement Applied to DNAzymes Confined to DNA Condensates
The goal of this study was to use toehold-mediated strand displacement to prevent or slow down the DNAzyme-catalyzed peroxidation reaction. The assumption was that the complementary displacer strand would bind to the nanostar, facilitated by the toeholds, and disrupt or unwind the G4 structure through displacement. Visually, this process should have translated to a clear or lighter blue color of the reaction tube in comparison to the positive control tube (without displacer strands). The displacer strand was added to the reaction tubes at 2X, 5X, and 10X concentrations (compared to the concentration of strand 4 of the nanostar with 5xG3T and a toehold attached), and the same amount of water was added to the positive control tube to account for the additional volume of the displacer strands.
Upon performing peroxidation experiments on NS15 DNA condensates with 5xG3T, and with hemin and 2X and 5X concentrations of the displacer strand added before and after annealing, it was observed that the only instances of prevention of the DNAzyme-catalyzed peroxidation reaction occurred when hemin was added post-annealing and the antis/displacer strands were added pre-annealing (Figure 5). The outcome of the pre-hemin experiments can be attributed to the thermodynamic stability of the G4/hemin complex, which is thermodynamically favored over toehold-mediated strand displacement. The G4 quadruplex forms through intramolecular interactions between guanine bases and is further stabilized by the potassium ion; on the other hand, TMSD requires intermolecular association between strands, and even at high concentrations of displacer strands with toeholds, it does not seem as though it is able to unwind the stable G4 structure. There is literature precedence for the stability of G4 quadruplexes [45],[46], and it is well known that the potassium cation stabilizes the G4 structure [24]. Although TMSD can be an incredibly quick reaction pathway [18],[19],[32],[33], it is also possible for this reaction to reverse, albeit at orders of magnitude slower rates [32]. In contrast, G4/hemin complexes are known to be stable once formed and resistant to structural rearrangements despite their slower, minute time scale needed for formation [47]. It is thus plausible that the toehold-mediated strand displacement reaction that occurs in the pre-hemin experiments is being reversed over the longer reaction period of 2 days, allowing for G4/hemin binding to take place, forming the active G4/hemin DNAzyme that can catalyze peroxidation when the substrates are added. To confirm these hypotheses, however, additional experiments must be conducted to measure the kinetics of toehold-mediated strand displacement on G-quadruplex structures and to compare these findings with the kinetics of G4/hemin complexing. It should further be noted that the pre-hemin experiment was also conducted with 10X concentration of displacer strand added pre-annealing with the same result (Figure S1). Similarly, we believe that adding the displacer strand after annealing the nanostar/forming the G4 quadruplex—after peroxidation has already begun—makes it difficult for TMSD to occur (even when hemin is added post-annealing), as it is not able to unwind the thermodynamically stable G4 quadruplex. Taken together, this data suggests that the G4/hemin complex formation is the favored and more stable outcome, and prevention of DNAzyme-catalyzed peroxidation reaction through TMSD requires substantial time (anti/displacer strand added pre-annealing) before the addition of the hemin co-factor, which binds to the G4 structure and yields a functional DNAzyme with peroxidase activity.
When NS15 DNA condensates with 5xG3T / toeholds attached to arm 4 were annealed with 2X and 5X concentrations of the corresponding antis/displacer strands, then incubated with hemin and peroxidized, both solutions were mostly clear with small pellets of blue color on the bottom of the tubes (Figure 5). This was hypothesized to be due to the hemin settling down and outcompeting the displacer strand to bind to the DNAzymes there, catalyzing the peroxidation. So, this procedure was repeated with the 5X anti/displacer tube, which contained 5X displacer strand, but the tube was flicked prior to peroxidation to resuspend the hemin, which resulted in an even clearer solution with only small wisps of blue throughout (Figure 6).
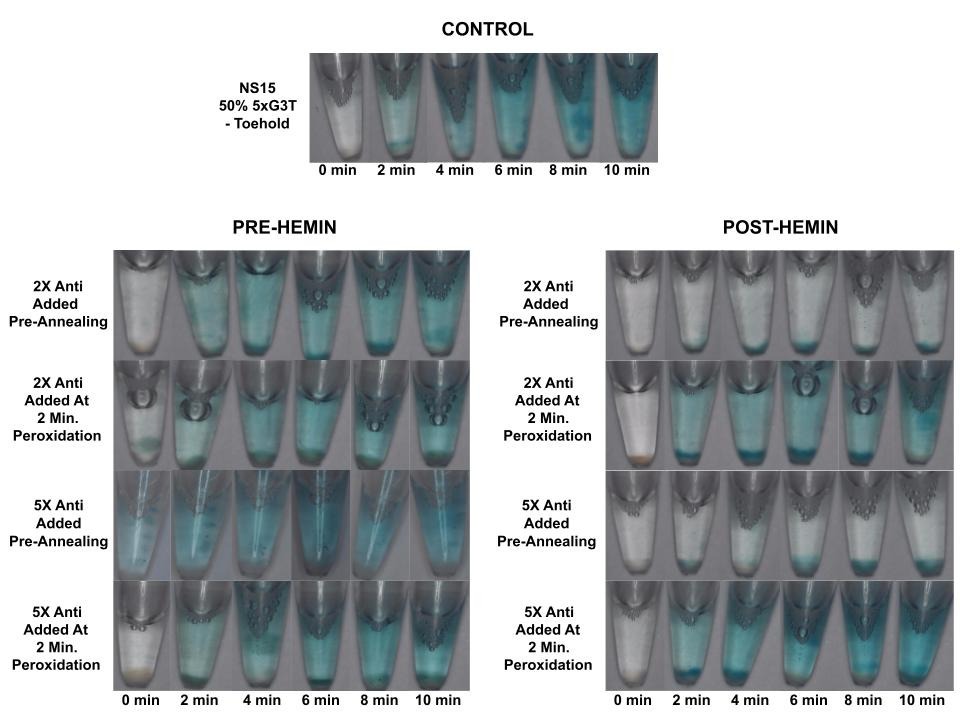
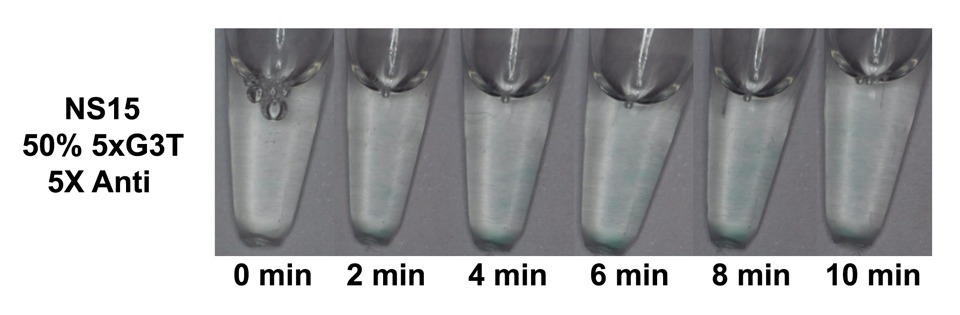
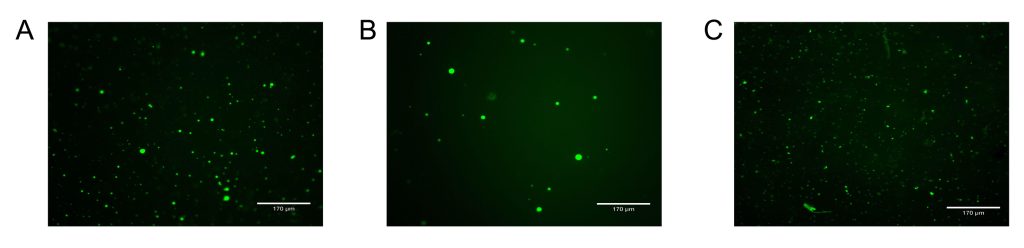
When a toehold was incorporated into the NS15 strand 4/G4 sequence and mixed with the corresponding displacer strands/toehold at 5X concentration prior to annealing, the anti/displacer strands appeared to largely prevent peroxidation when hemin was added post-annealing (Figure 6). On the other hand, when the same concentration of anti/displacer strand was added post-annealing, the solution appeared as blue as the control tube, hypothesized to be due to the anti/displacer strand having less time and/or opportunity to compete with the formation of the G4/hemin complex when it was added after hemin incubation and once the peroxidation reaction had already been initiated (Figure 5). Interestingly, when samples from the NS15/50% 5xG3T/toehold reaction tubes were viewed under an Echo Revolve Fluorescence Microscope to ensure condensate formation, it was observed that DNA condensate formation was reduced on the microscale when toeholds and anti/displacer strands were added in comparison to the positive control (Figure 7). We have observed a similar phenomenon on the macroscale while forming bulk layers of orthogonal DNA condensates; when adding anti/displacer strands complementary to nanostar strand 4/G4 prior to annealing, a DNA condensate layer was not observed [48].
Conclusions
The strategy of toehold-mediated strand displacement was explored as a means of disrupting the formation of the G4/hemin complex, which would prevent or slow down the peroxidation reaction it catalyzes. Although this strategy yielded mixed results, it led to new research ideas regarding the stability of the G4 quadruplex structure and whether TMSD could be used to displace or unravel the DNAzyme in a robust and tunable manner. Ideally, the reversibility of TMSD should enable programming of the DNAzyme-mediated peroxidation reaction and the ability to turn the reaction on and off within synthetic DNA condensates. These experiments show that TMSD may be a viable method of reducing, if not preventing, the progress of the DNAzyme-catalyzed peroxidation reaction, but further fine-tuning of this strategy may be necessary; perhaps longer toeholds can be used in TMSD design strategies. Different G4 quadruplexes, such as those explored in previous work localizing DNAzyme-catalyzed peroxidation to DNA condensates [30] can also be used in studies attempting to use TMSD to program DNAzyme-catalyzed peroxidation.
When 5X concentration of displacer/anti strands were added prior to annealing, incubated with hemin after annealing, and then peroxidized, most of the solution was clear with blue pellets when unshaken prior to peroxidation (Figure 5) and with a few wisps of blue when shaken (Figure 6). The best result was achieved under the latter conditions, and the 5X anti/displacer strands were hypothesized to have prevented folding of the G4 quadruplex, with the exception of a few that may have initiated peroxidation. For instance, there were small blue “droplets” within the solution, which may have been a result of DNAzyme complexes catalyzing peroxidation, but this is speculative and requires further investigation.
Since this research was conducted through the PCC nanostar undergraduate research program that takes place over six weeks every summer, 20 hours per week, there are some unanswered questions that remain to be explored regarding aspects of these experimental results. As done by every previous cohort of the nanostar program thus far, we leave behind suggestions for future exploration of toehold-mediated strand displacement for the 2025 summer cohort—a new group of diverse and underrepresented undergraduate researchers who will be given the unique opportunity to be trained by graduate students at UCLA and to investigate these remaining questions firsthand. In future experiments, we suggest that tubes with 2X and 10X concentrations of anti/displacer strands are incubated with hemin after annealing and shaken up by hand prior to peroxidation to compare results with the 5X reaction tubes in Figure 6. Further, the same experiments investigated in this study should be explored using different DNAzymes and DNA nanostars with greater than 15 base pairs per arm to determine if similar results are observed; this is important because the structure of each unique DNAzyme results in varying degrees of stability [46],[47],[49], and some DNAzymes (e.g., LA4), result in a stronger color change—even when localized to DNA condensates [30].
Although promising results were achieved with 5X concentration of anti/displacers strands, TMSD does not seem to be the most reliable, nor the most effective, method to prevent this DNAzyme-catalyzed peroxidation reaction. In the future, different methods of unwinding, or disrupting, the G4 quadruplex to ‘turn off’ the DNAzyme-catalyzed peroxidation reaction should be explored, such as through the use of G4 helicases and/or small-molecule G4 destabilizers [34]. The ABTS radical cation reaction product can be monitored through spectroscopy by measuring the absorbance at 420 nm, as done in this study.
PCC nanostar researchers presented this scientific research at PCC and at the ACS National Meeting in 2025, and aspects of this work were taught in Dr. J.B.’s General, Organic, and Biochemistry course and at Girls Science Day (Tech Savvy) at PCC, inspiring a diverse body of students with DNA nanotechnology. This novel synthetic biology curriculum represents a paradigm shift in STEM education, as it provides an open-ended platform for students to do authentic research in a course-based setting and apply their knowledge through active learning. This exemplifies the meta-learning framework [50], systems thinking in chemistry education [51], and by making YouTube tutorials that teach students how to simulate nanostars [52] and conduct other aspects of this research, we are creating an accessible, engaging, and global platform for STEM education focused on emerging technologies to inspire students to imagine & build a sustainable, innovative future.
Acknowledgments. We would like to express immense gratitude to our collaborators, Deborah Fygenson at UC Santa Barbara and Paul Rothemund at Caltech, for their unwavering support and guidance throughout this research program. We would also like to thank the graduate students and postdocs in Elisa Franco’s laboratory at UCLA, Deborah Fygenson’s laboratory at UCSB, and Paul Rothemund’s laboratory at Caltech for their guidance, mentorship and support at the UCLA research techniques bootcamp. We thank the students in Dr. J.B.’s General, Organic and Biochemistry course at PCC in the Spring 2023–2025 semesters for their enthusiastic participation in the DNA nanotechnology laboratories based on this research, as well as the young girls who participated in Girls Science Day in 2023–2025. We acknowledge NSF FMRG: Bio Award 2134772 for funding of this research, education, and workforce development program.
Disclosures. The authors declare no conflicts of interest.
Supplemental Materials:
[1] Seeman, N.C. & Sleiman, H.F., “DNA nanotechnology.” Nature Reviews Materials, vol. 3, 2018.
[2] Bathe, M. & Rothemund, P.W.K., “DNA Nanotechnology: A Foundation for Programmable Nanoscale Materials.” MRS Bulletin,vol. 42, 882–888, 2017.
[3] Lafontaine, D.L.J., Riback, J.A., Bascetin, R. & Brangwynne, C.P., “The nucleolus as a multiphase liquid condensate.” Nat. Rev. Mol. Cell Biol. vol. 22, 165–182, 2021.
[4] Kumar, K., Mella-Herrera, R.A. and Golden, J.W., “Cyanobacterial heterocysts,” Cold Spring Harb. Perspect. Biol., vol. 2, no. 4, Apr. 2010, doi: 10.1101/cshperspect.a000315.
[5] Zwicker, D., Decker, M., Jaensch, S., Hyman, A.A. & Jülicher, F., “Centrosomes are autocatalytic droplets of pericentriolar material organized by centrioles.” Proc. Natl. Acad. Sci. U.S.A. vol. 111, E2636–45, 2014.
[6] Hyman, A. A., Weber, C. A. & Jülicher, F., “Liquid-liquid phase separation in biology.” Annu. Rev. Cell Dev. Biol. vol. 30, 39–58, 2014.
[7] Banani, S.F., Lee, H. O., Hyman, A.A. & Rosen, M. K., “Biomolecular condensates: organizers of cellular biochemistry.” Nat. Rev. Mol. Cell Biol. vol. 18, 285–298, 2017.
[8] O’Flynn, B. G. & Mittag, T., “The role of liquid–liquid phase separation in regulating enzyme activity.” Current Opinion in Cell Biology, vol. 69, 70–79, 2021.
[9] Feric, M. et al., “Coexisting Liquid Phases Underlie Nucleolar Subcompartments.” Cell, vol. 165, 1686–1697, 2016.
[10] Le Feuvre, R.A. & Scrutton, N.S., “A living foundry for Synthetic Biological Materials: A synthetic biology roadmap to new advanced materials.” Synth. Syst. Biotechnol., vol. 3, 105–112, 2018.
[11] Bueso, Y.F. & Tangney, M., “Synthetic Biology in the Driving Seat of the Bioeconomy.” Trends in Biotechnology, vol. 35, 373–378, 2017.
[12] Bracha, D., Walls, M.T. & Brangwynne, C.P., “Probing and engineering liquid-phase organelles,” Nat. Biotechnol., vol. 37, no. 12, pp. 1435-1445, Dec. 2019, doi: 10.1038/s41587-019-0341-6.
[13] Jeon, B.J., Nguyen, D.T. & Saleh, O.A., “Sequence-controlled adhesion and microemulsification in a two-phase system of DNA liquid droplets,” J. Phys. Chem. B., vol. 124, no. 40, pp. 8888-8895, Sep. 2020, doi: 10.1021/acs.jpcb.0c06911.
[14] Kojima T. & Takayama, S., “Membraneless compartmentalization facilitates enzymatic cascade reactions and reduces substrate inhibition,” ACS Appl. Mater. Interfaces, vol. 10, no. 38, pp. 32782-32791, Sep. 2018, doi: 10.1021/acsami.8b07573.
[15] Reinkemeier, C.D., Girona, G.E. & Lemke, E.A., “Designer membraneless organelles enable codon reassignment of selected mRNAs in eukaryotes.” Science, vol. 363, 2019.
[16] Sato, Y., Sakamoto, T.; Takinoue, M., “Sequence-based engineering of dynamic functions of micrometer-sized DNA droplets”, Sci. Adv. vol. 6: eaba3471, 2020.
[17] Do, S., Lee, C., Lee, T., Kim, D-N., Shin, Y., “Engineering DNA-based synthetic condensates with programmable material properties, compositions, and functionalities.” Sci. Adv., vol. 8, eabj1771, 2022.
[18] Simmel, F.C., Yurke, B. & Singh, H.R., “Principles and Applications of Nucleic Acid Strand Displacement Reactions.” Chem. Rev., vol. 119, 6326–6369, 2019.
[19] Zhang, D.Y. & Seelig, G., “Dynamic DNA nanotechnology using strand-displacement reactions,” Nat. Chem., vol. 3, no. 2, pp. 103-113, Feb. 2011, doi: 10.1038/nchem.957.
[20] Blatti, J.L., “Invited Letter: Undergraduate research as a means to build a creative, resilient, and highly skilled biomanufacturing workforce.” J. Adv. Tech. Educ. 2023, vol. 2, no. 2, doi: https://doi.org/10.5281/zenodo.8342997.
[21] Biffi, S. et al., “Phase behavior and critical activated dynamics of limited-valence DNA nanostars,” Proc. Natl. Acad. Sci.U.S.A. vol. 110, no. 39, pp. 15633-15637, Sep. 2013, doi: 10.1073/pnas.1304632110.
[22] Conrad, N., Kennedy, T., Fygenson, D.K. and Saleh, O.A., “Increasing valence pushes DNA nanostar networks to the isostatic point,” PNAS, vol. 116, no. 15, pp. 7238-7243, Mar. 2019, doi: 10.1073/pnas.1819683116.
[23] Agarwal, S., Osmanovic, D., Dizani, M., Klocke, M. A. and Franco, E., “Dynamic control of DNA condensation,” Nat Commun, vol. 15, no. 1915, Mar. 2024, doi: 10.1038/s41467-024-46266-z.
[24] Travascio, P., Li, Y. & Sen, D., “DNA-enhanced peroxidase activity of a DNA-aptamer-hemin complex.” Chem. Biol. vol. 5, pp. 505–517, 1998.
[25] Adeoye, R.I. et al., “Catalytic activities of multimeric G-quadruplex DNAzymes,” Catal., vol. 9, no. 7, p. 613, Jul. 2019, doi: 10.3390/catal9070613.
[26] Stefan, L., Xu, H-J., Gros, C.P., Denat, F., Monchaud, D., “Harnessing Nature’s Insights: Synthetic Small Molecules with Peroxidase-Mimicking DNAzyme Properties.” Chem. Eur. J., vol. 17, pp. 10857-10862, 2011, doi: https://doi.org/10.1002/chem.201101337.
[27] Chen, J., Zhang, Y., Cheng, M., Guo, Y., Šponer, J., Monchaud, D., Mergny, J.L., Ju, H., Zhou, J., “How Proximal Nucleobases Regulate the Catalytic Activity of G-Quadruplex/Hemin DNAzymes”, ACS Catal., vol. 8, no. 12, pp. 11352-11361, 2018.
[28] Li, W., Li, Y., Liu, Z., Lin, B., Yi, H., Xu, F., Nie, Z., Yao, S., “Insight into G-quadruplex-hemin DNAzyme/RNAzyme: adjacent adenine as the intramolecular species for remarkable enhancement of enzymatic activity”, Nucleic Acids Res., vol. 44, no. 15, pp. 7373-84, 2016.
[29] Kosman, J., Żukowski, K. & Juskowiak, B., “Comparison of Characteristics and DNAzyme Activity of G4–Hemin Conjugates Obtained via Two Hemin Attachment Methods.” Molecules: A Journal of Synthetic Chemistry and Natural Product Chemistry, vol. 23, 2018.
[30] Anand, T., Salman, R., Castaneda-Camacho, B., Gonzalez, V., Safar, E., Franco, E., Blatti, J.L., “Investigating substrates Amplifu Red and ABTS (2,2’-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) in the colorimetric detection of DNAzyme activity localized to DNA condensates,” J. Adv. Technol. Educ. vol. 4, no. 1, 2025, doi: 10.5281/zenodo.14933441. Figure 2 Created in BioRender. Blatti, J. (2025) https://BioRender.com/pfeb4um.
[31] Terkazaryan, E., Salman, R., Paredes, F., Gonzalez, V., Safar, E., Yu, W.L., Mhlanga, G., Rivera, M., Khosravian, C., Pithawalla, D., Anand, T., Castaneda-Camacho, B., Romero Mercieca, H., Dizani, M., Franco, E., Blatti, J.L., “Molecular visualization in nanotechnology education, research, and outreach: simulating catalytic DNA nanostars using oxDNA.” J. Adv. Technol. Educ., 2025, vol. 5, no. 1, doi: https://doi.org/10.5281/zenodo.17041761.
[32] Zhang, D.Y. & Winfree, E., “Control of DNA strand displacement kinetics using toehold exchange,” J. Am. Chem. Soc., vol. 131, no. 47, pp. 17303-17314, 2009, doi: 10.1021/ja906987s.
[33] Srinivas, N. et al., “On the biophysics and kinetics of toehold-mediated DNA strand displacement,” Nucleic Acids Res., vol. 41, no. 22, pp. 10641-10658, Dec. 2013, doi: 10.1093/nar/gkt801.
[34] Lejault, P., Mitteaux, J., Sperti, F.R. & Monchaud, D., “How to untie G-quadruplex knots and why?” Cell Chem. Biol., vol. 28, no. 4, pp. 436-455, Apr. 2021, doi: 10.1016/j.chembiol.2021.01.015.
[35] Terkazaryan, E. (2025) https://BioRender.com/h84a388.
[36] Tanaka, S., Chan, S.K., Lim, T.S., Ohya, Y. & Kuzuya, A., “Communication—DNA quadruplex hydrogel beads showing peroxidase activity,” J Electrochem. Soc., vol. 166, no. 9, pp. B3271-B3273, May 2019, doi: 10.1149/2.0441909jes.
[37] Drobot, B. et al., “Compartmentalised RNA catalysis in membrane-free coacervate protocells.” Nat. Commun. vol. 9, pp. 3643, 2018.
[38] Thomas, I.B.K., Gaminda, K.A.P., Jayasinghe, C.D., Abeysinghe, D.T., Senthilnithy, R., “DNAzymes, Novel Therapeutic Agents in Cancer Therapy: A Review of Concepts to Applications.” J. Nucleic Acids. Nov 1, 2021:9365081. doi: 10.1155/2021/9365081.
[39] Christopher Galicia, “The ‘Nanostars’ of PCC’s synthetic biology research program”, PCC Courier, Sept. 11, 2024.
[40] Christopher Galicia, “Posters of Nanostar research break down the hard science behind research breakthroughs”, PCC Courier, Oct. 24, 2024.
[41] Ashcroft, J.M., Blatti, J.L., Jaramillo, V.J., “Early Career Undergraduate Research as a Meaningful Academic Experience in which Students Develop Professional Workforce Skills: A Community College Perspective.” In: Becoming a Chemist: Scaffolding Professional Skills into Undergraduate Curricula; Neiles, K.Y.; Mertz, P.S.; and Fair, J.D. Eds.; ACS Symposium Series, vol. 1365; American Chemical Society: Washington DC, 2020; pp 281-299.
[42] Hernandez, P. R., Woodcock, A., Estrada, M. & Wesley Schultz, P., “Undergraduate Research Experiences Broaden Diversity in the Scientific Workforce.” BioScience vol. 68, 204–211, 2018.
[43] Zadeh, J. N. et al., “NUPACK: Analysis and design of nucleic acid systems.” J. Comput. Chem. vol. 32, pp. 170–173, 2011.
[44] EDVOTEK | The Biotechnology Education Company. “Quick Guide: SYBR® Safe DNA Stain.” Accessed: Dec. 14, 2024. [Online.] Available: https://www.edvotek.com/quick-guide-sybr-safe.
[45] Jana, J. & Weisz, K., “Thermodynamic Stability of G-Quadruplexes: Impact of Sequence and Environment.” ChemBioChem, vol. 22, pp. 2821-2907, 2021.
[46] Nakata, M., Kosaka, N., Kawauchi, K., & Miyoshi, D., “Quantitative Effects of the Loop Region on Topology, Thermodynamics, and Cation Binding of DNA G-quadruplexes.” ACS Omega vol. 9 (32), pp. 35028-35036, 2024, doi: 10.1021/acsomega.4c05008.
[47] Gui, Y., Chen, J., Cheng, M., Monchaud, D., Zhou, J, Ju, H., “A Thermophilic Tetramolecular G-Quadruplex/Hemin DNAzyme.” Ang. Chemie, vol. 56 (52), pp. 16636-16640, 2017.
[48] unpublished results
[49] Bochman, M., Paeschke, K., & Zakian, V., “DNA secondary structures: stability and function of G-quadruplex structures,” Nat Rev Genet, vol. 13, pp. 770-780, Oct. 2012, doi: 10.1038/nrg3296.
[50] Fadel, C., 2019. Business at OECD (BIAC) Position Paper for the proposed International Symposium on Employability and the Learner Profile (ISELP), Center for Curriculum Redesign, Massachusetts Institute of Technology, Retrieved November 15, 2019, from https://curriculumredesign.org/ wp-content/uploads/EDU-EDPC-RD-2019-14-Declassified.pdf.
[51] Blatti, J.L. et al., “Systems Thinking in Science Education and Outreach toward a Sustainable Future.” J. Chem. Educ., vol. 96, pp. 2852–2862, 2019.
[52] Paredes, Fabiana., “Simulating DNA Nanostars Using oxDNA and oxView: Tutorial.” YouTube, 15 Aug. 2024, youtu.be/_FrL3UHvhdA.